This was the origin and motivation for our project “2Detect – A large 2D expandable, trainable, experimental Computed Tomography data collection for machine learning”, which aimed to acquire a large 2D computed tomography dataset for a broad range of application areas, such as denoising, sparse- and limited-angle scanning, beam-hardening reduction, super-resolution, region-of-interest tomography or segmentation. However, there are several requirements involved in acquiring such a large 2D CT dataset. Firstly, research groups must have easy access to a scanning facility, which can be used for a time-consuming data collection process on a large scale. Secondly, the employed scanner must have the capability to adjust the acquisition geometry and acquisition parameters to ensure that the dataset collected is suitable for a wide range of machine learning applications. Thirdly, the image characteristics should closely resemble those encountered in medical CT, as medical imaging is a crucial application area for X-ray CT. Lastly, it is essential to minimize manual intervention during the acquisition process in order to obtain a substantial number of CT reconstruction slices. This necessitates automating the acquisition process as much as possible.
Since our “Computational Imaging” research group at Centrum Wiskunde & Informatica (CWI) could meet all these requirements, we took on the challenge to create such a large 2D Computed Tomography dataset. For this, it was especially important to understand the needs of the community and the prerequisites on the data we wanted to collect. Furthermore, we needed to acquire an in-depth knowledge about all the capabilities of the FleX-ray scanner, a highly flexible, programmable and custom-built X-ray CT scanner located at CWI. This was integral to the selection and testing of the various acquisition parameters and scanning objects for achieving versatile application areas for the dataset, such as denoising, sparse- and limited-angle scanning, beam-hardening reduction, super-resolution, region-of-interest tomography or segmentation.
Objectives
The objectives were to acquire at least a noisy, a beam-hardening artifact-inflicted and a “clean” scan as a ground truth and starting point for constructing a high-confidence segmentation, since data for sparse- and limited-angle, super-resolution and region-of-interest tomography can be generated from scans with a large amount of angle projections and high resolution. Therefore, data had to be acquired in three different acquisition modes: Mode 1: the noisy, low-dose acquisition; mode 2: the clean, high-dose acquisition; and mode 3: the beam hardening artifact-inflicted acquisition. Choosing suitable acquisition parameters for these different modes was a difficult process and required a lot of test scans.
The pressure on making “the right choices” for such a large-scale project was quite big, because we wanted to be certain that these choices will actually enable the versatile application areas and that every consideration will work in the end. Therefore, the first nine months after the initial idea of the dataset were used for meticulous testing of potential sample objects, potential acquisition parameters and potential scanning protocols. This included finding suitable sample containers by browsing in hardware and craft stores and testing their properties in the CT scanner. It also involved upgrading the capabilities of the FleX-ray scanners since the device did not contain the possibility to filter the X-ray beam used for acquiring the CT scans. The author had to design and 3D-print an extension to the FleX-ray scanner that would enable the placement of metal filter sheets of varying materials and thicknesses.
Visiting a garden centre
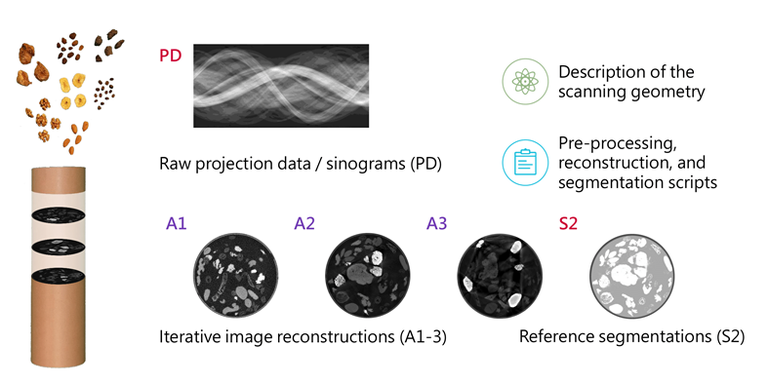
Furthermore, we searched for a suitable mix of samples with high natural variability in shape and density, resembling the image characteristics found in abdominal CT scans. This encompassed buying various food items from the supermarket and scanning all kinds of fruits, vegetables, and nuts to evaluate their appearance in CT scans. The final mix consisted of almonds, dried banana chips, coffee beans, dried figs, raisins, and walnuts immersed in cereal-based coffee powder as a filler material (cf. Fig. 2). Unfortunately, the image reconstructions of this sample mix showed no visual differences between the different acquisition modes selected earlier.



![FleX-ray Lab: the computed tomography set-up used for the data acquisition. (1) Cone-beam X-ray source; (2) Thoraeus filter sail [Sn 0.1mm, Cu 0.2mm, Al 0.5mm]; (3) Rotation stage; (4) Sample tube; (5) Flat panel detector. The objects 1, 3, 4, and 5 move from their red transparent front position to the mid position for the acquisitions of mode 3. In both positions 3,601 projection images per slice are taken while the object rotates 360 degrees.](/images/44402/maxblog4.width-768.jpg)

