Our team, which is led by Solon Pissis, works on algorithms and data structures for sequence analysis.
Research topics we are working on include algorithms and data structures on strings for pattern matching, indexing, comparison, and finding regularities. These topics are collectively known as combinatorial pattern matching. Combinatorial pattern matching methods are typically the workhorse of computational molecular biology applications, where the raw data to be analyzed are DNA, RNA or protein sequences. We are interested in such applications but also in other applications processing sequential data, such as data mining, data compression or information retrieval applications.
Combinatorial pattern matching
Let us start by providing a brief overview of the combinatorial pattern matching topics.
In pattern matching, we are given a string x to pre-process in time that is proportional to the length of x, so that when we are given another longer string y, we can find all occurrences of x in y in time that is proportional to the length of y. In the following example we see two occurrences of string x in string y.
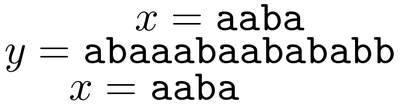
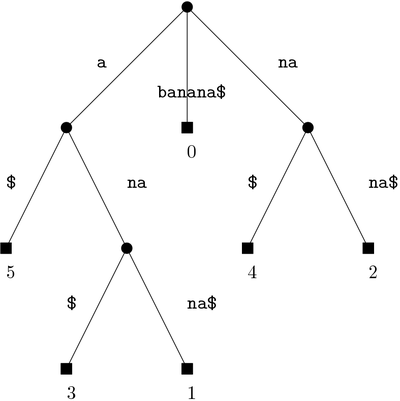
In indexing, we are given a string y to pre-process into a data structure in time that is proportional to the length of y, so that when we are given another shorter string x, we can find occurrences of x in y in time that is proportional to the length of x. In the following example we see an indexing data structure for string y = banana$. Every path from the root to a leaf node corresponds to a suffix of y. For instance, nana$ is the suffix starting at position 2 of y using a 0-base index.
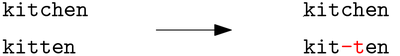
In sequence comparison, we are given two (or more) strings and the task is to compare them in order to infer their (dis)similarities. In the following example we see an alignment of two strings: kitchen and kitten.
In finding regularities, we are given a string x and the task is to find certain types of patterns repeating in x. In the following example we see that pattern aabbab repeats in string x = abaabbabbaaabbabbba.
Note the fundamental difference to pattern matching: here, the pattern is not given to us but we are rather asked to extract it from x.
Computational biology
Let us mention a few applications of combinatorial pattern matching in computational molecular biology:
- The first step of genome assembly via read mapping is to construct an indexing data structure over the reference genome. The first step of de novo genome assembly is to find common substrings between the reads to construct a de Bruijn or an overlap graph.
- The first step of inferring evolutionary relationships between sequences is the pairwise or multiple comparison of these sequences. The output of this step is used as the input to methods that reconstruct phylogenetic trees to depict these relationships.
- Inferring functional relationships between sequences is performed via comparing a sequence to a sequence database to detect regions of significant similarity or via extracting patterns (motifs), which are significantly overrepresented in a set of sequences.
Data mining
Combinatorial pattern matching methods are also applied in data mining, when the data to be analyzed are textual (sequential) and unstructured. For example, a string can represent the movement history of an individual, with each letter corresponding to a visited location; or an individual's purchasing history in a retailer, with each letter corresponding to a purchased product. Let us mention a few such applications:
- In pattern mining the task is to extract actionable patterns from large textual datasets; e.g. to extract the most frequent or the most significant patterns.
- In data sanitization the task is to disguise confidential patterns in large textual datasets while preserving the utility of these datasets.
- In document clustering the task is to compute term frequencies to reflect how important a term is to a document in a large collection of documents.
Researchers
Current projects with external funding
- MSCA-RISE-2019 - Research and Innovation Staff Exchange (PANGAIA)
- Royal Society International Exchanges 2023 with the University of London
Recent related research outputs
- Lorraine A. K. Ayad, Grigorios Loukides, Solon P. Pissis: Text indexing for long patterns using locally consistent anchors. The VLDB Journal, 2025.
- Ben Bals, Sebastiaan van Krieken, Solon Pissis, Leen Stougie, Hilde Verbeek. When is String Reconstruction using de Bruijn Graphs Hard? ESA 2025
- Giulia Bernardini, Huiping Chen, Alessio Conte, Roberto Grossi, Veronica Guerrini, Grigorios Loukides, Nadia Pisanti, Solon P. Pissis: Indexing Strings with Utilities. ICDE 2025
- Solon P. Pissis: Optimal prefix-suffix queries with applications. SOSA 2025
- Estéban Gabory, Chang Liu, Grigorios Loukides, Solon P. Pissis, Wiktor Zuba: Space-Efficient Indexes for Uncertain Strings. ICDE 2024
Alumni
- Wiktor Zuba (Postdoc, Mar 2022 - Aug 2024)
- Esteban Gabory (Ph.D student, Oct 2021 - Aug 2024)
- Michelle Sweering (Ph.D student, Nov 2019 - Jan 2024)
- Giulia Bernardini (Postdoc, Jan 2021 - Dec 2021)
Past projects with external funding
- MSCA-ITN-2020 - Innovative Training Networks (ALPACA)
