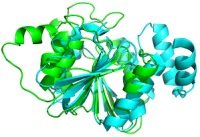
CWI researcher Inken Wohlers has developed a new method to compare protein shapes. This enables biologists to discover the function of new proteins more efficiently. She will defend her thesis on 11 December 2012. Wohlers developed mathematical models and computer algorithms to compare two protein structures optimally according to given criteria. Biologists compare proteins to determine the function of a protein from its structure. Protein molecules consist of tens to thousands of amino acids, folded into complex, 3-dimensional shapes. This structure determines the function of the protein. Therefore, biologists compare the structure of proteins whose function is unknown to those of known proteins. Comparing proteins is also instrumental in evolutionary biology: it shows the evolutionary relationship between certain families of proteins.
Because of their complex structure, comparing proteins accurately is difficult. Biologists use computer algorithms to do this, but there is no consensus over which method to use. There are many criteria that can be used to compare proteins, leading to a myriad of different methods that are currently in use. The general frameworks presented in Wohlers' thesis incorporate existing criteria as well as novel criteria and calculate an optimal solution based on that.
Thesis: Exact algorithms for pairwise protein structure alignment
By: Inken Wohlers (CWI Life Sciences-group)
Promotor: prof. dr. Gunnar Klau (CWI and VU)
Date: 11 December 2012, 13.45h
Place: VU Aula
See also:
CWI builds online application for protein comparison - 30 July 2012
